Next Generation Synthesis. Week 4
Important Notions
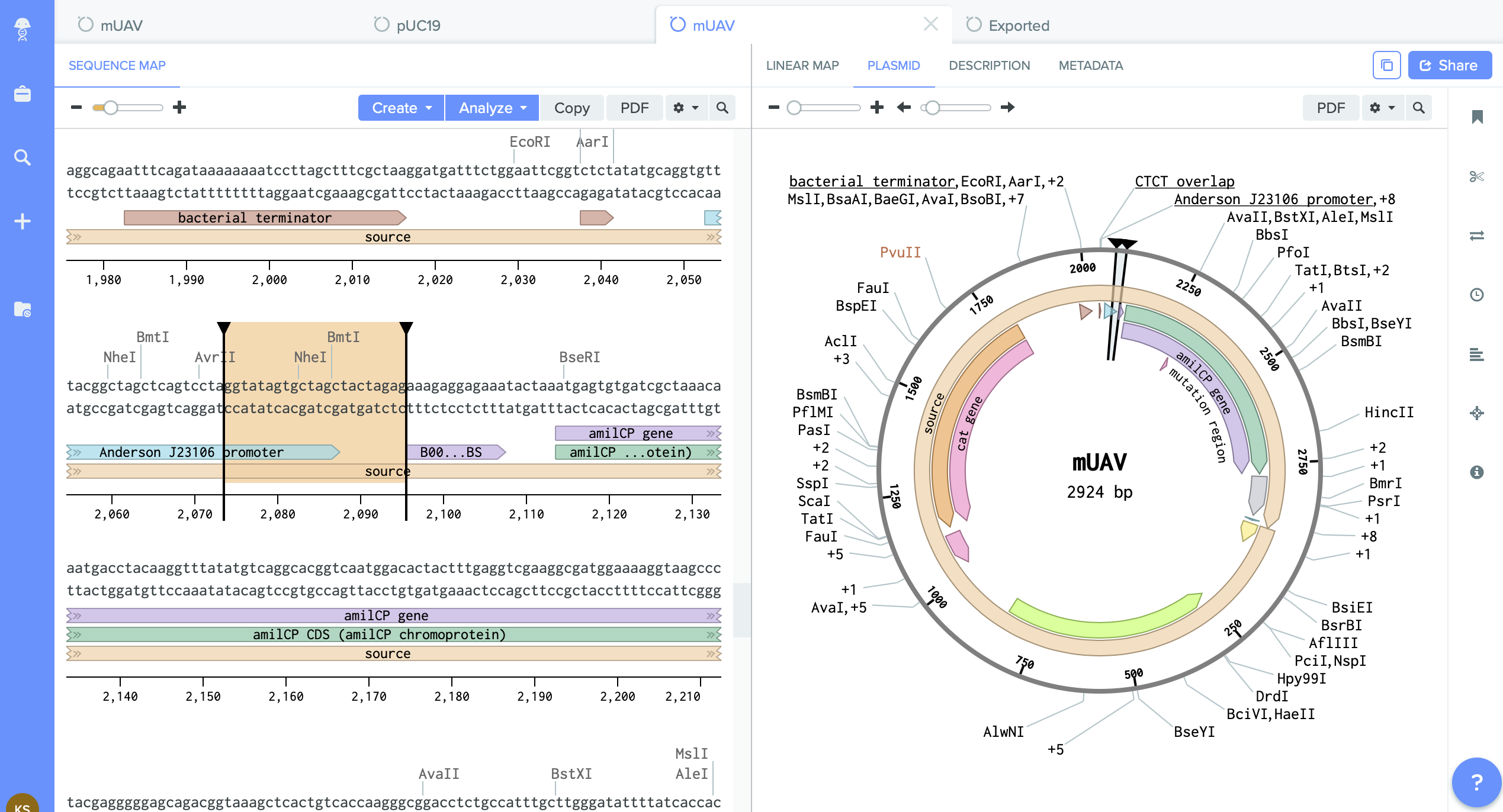
Designing Plasmids
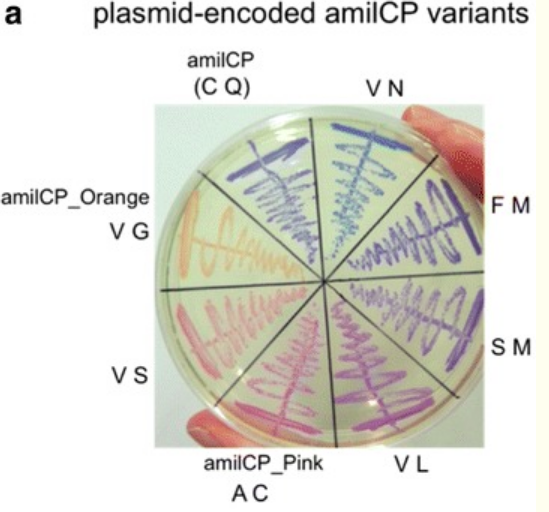
Amil CP variants
| Variant | amino acids | Bases |
|---|---|---|
| Original | C Q (Cysteine, Glutamine) | TGTCAG |
| Orange | V G (Valine, Glycine) | (GTT/GTC/GTA/GTG)+(GGT/GGC/GGA/GGG) |
| Light Pink | V S (Valine, Serine) | (GTT/GTC/GTA/GTG) + (TCT/TCC/TCA/TCG/AGT/AGC) |
| amilCP_Pink | A C (Alanine, Cysteine) | (GCT/GCC/GCA/GCG) + (TGT/TGC) |
| Darker Pink/Magenta | V L (Valine, Leucine) | (GTT/GTC/GTA/GTG) + (CTT/CTC/CTA/CTG/TTA/TTG) |
| Violet | S M (Serine, Methionine) | (TCT/TCC/TCA/TCG/AGT/AGC)+ ATG |
| Darker Violet | F M (Phenylalanine, Methionine) | (TTT/TTC)+ ATG |
| Bleu | V N (Valine, Asparagine) | (GTT/GTC/GTA/GTG) + (AAT/AAC) |
Important notions:
Promoter
- A promoter is a region of DNA where transcription of a gene is initiated. Promoters are a vital component of expression vectors because they control the binding of RNA polymerase to DNA. RNA polymerase transcribes DNA to mRNA which is ultimately translated into a functional protein. Thus the promoter region controls when and where in the organism your gene of interest is expressed.
RBS
- Ribosome binding site - is a sequence of nucleotides upstream of the start codon of an mRNA transcript that is responsible for the recruitment of a ribosome during the initiation of translation.
Difference between Promoter and RBS
- Promoter is the site where RNA polymerase binds to. This is related to transcription. Ribosomes binding site is where the ribosomes binds. This is related to translation. For eukaryotic mRNA, this occurs by the recognition 5 methyl group ( remember the mRNA processing for eukaryotes). For prokaryotes, the ribosome binding site is the shine dalgarno sequence
Terminator
For this assingment we are working with two plasmids.
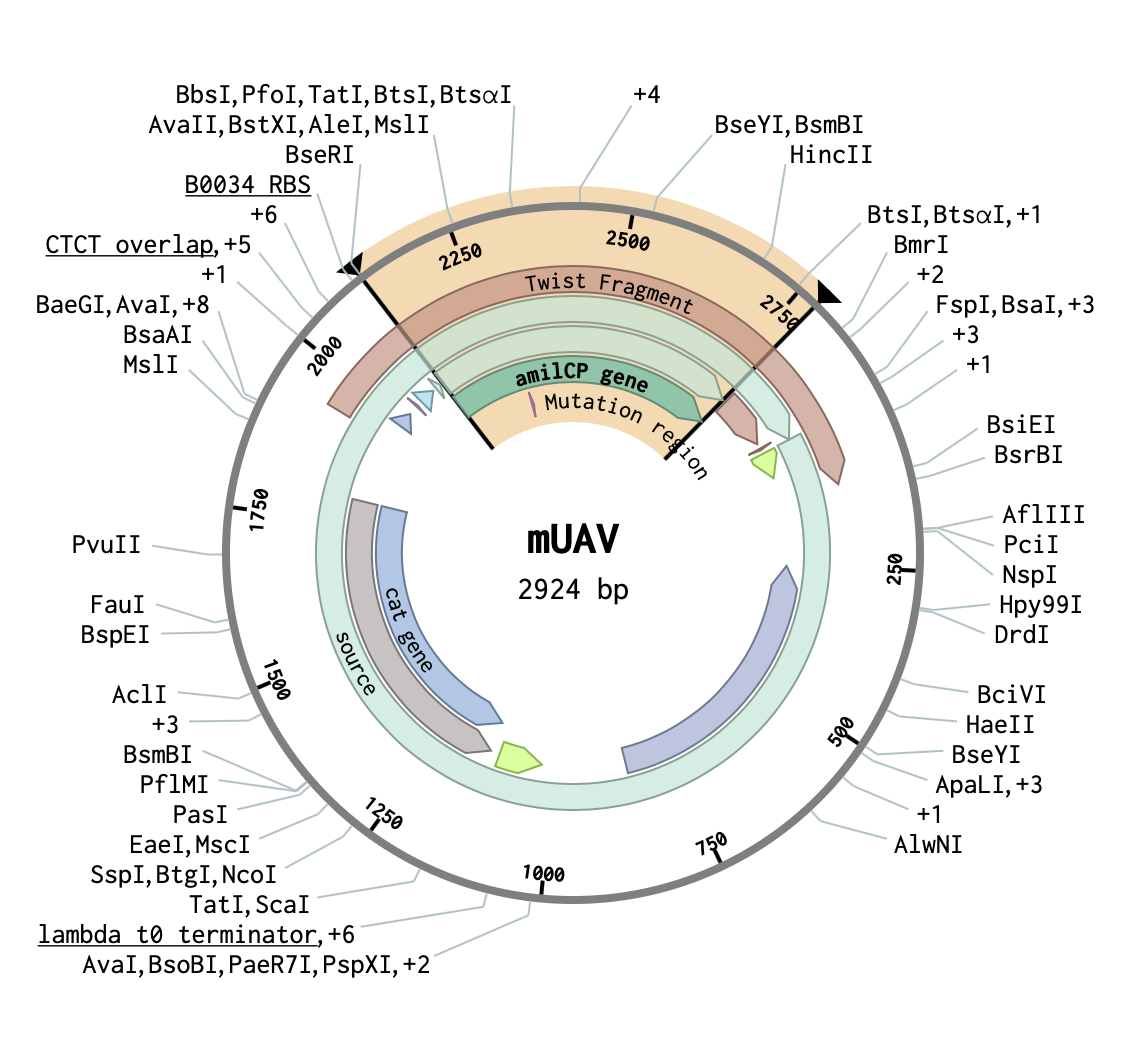
1) mUAV- Mobius Assembly Universal Acceptor Vector. (It converts amplified PCR fragments into standard exchangable parts.
We have on it amilCP gene which I want to mutate.
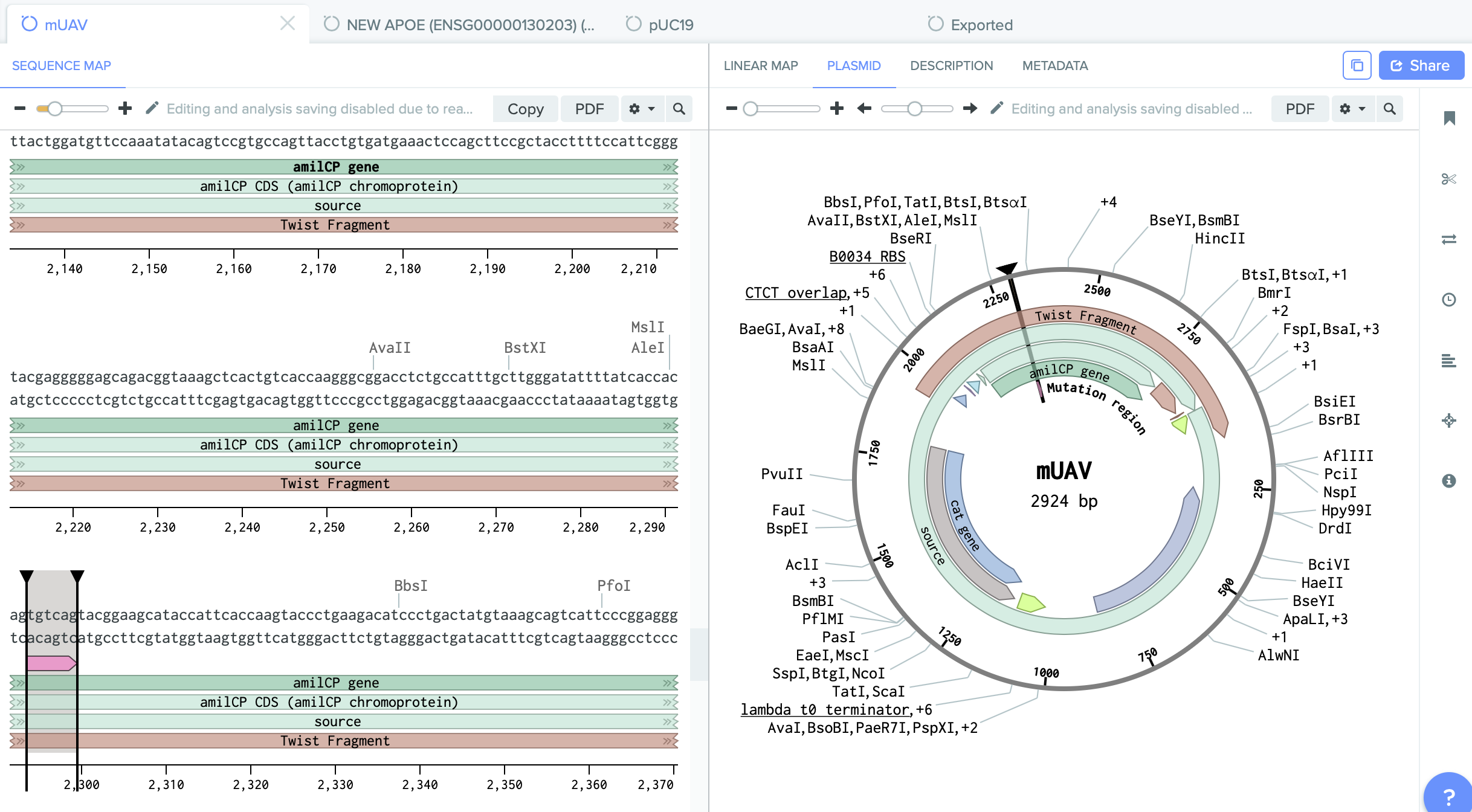
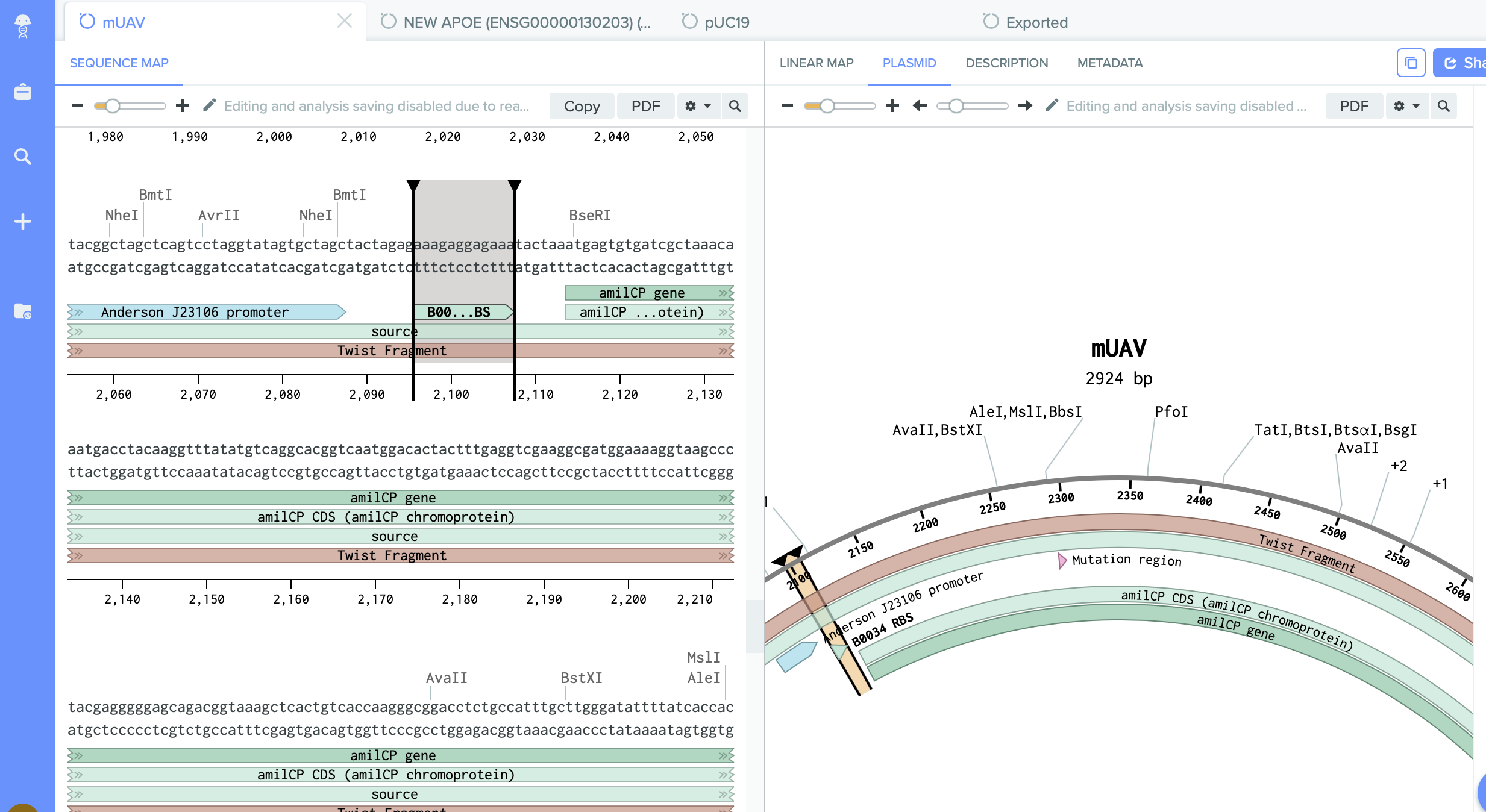
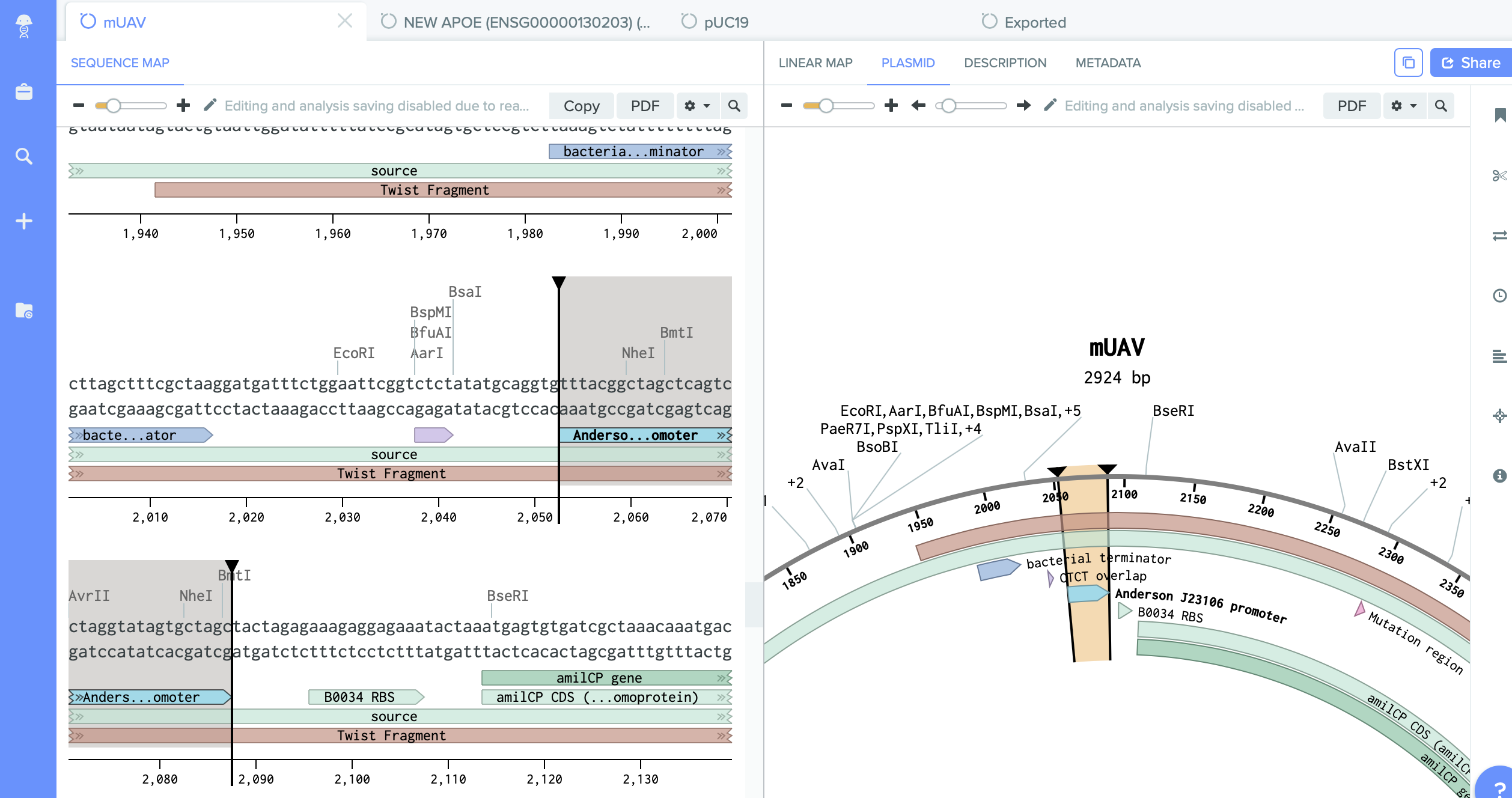
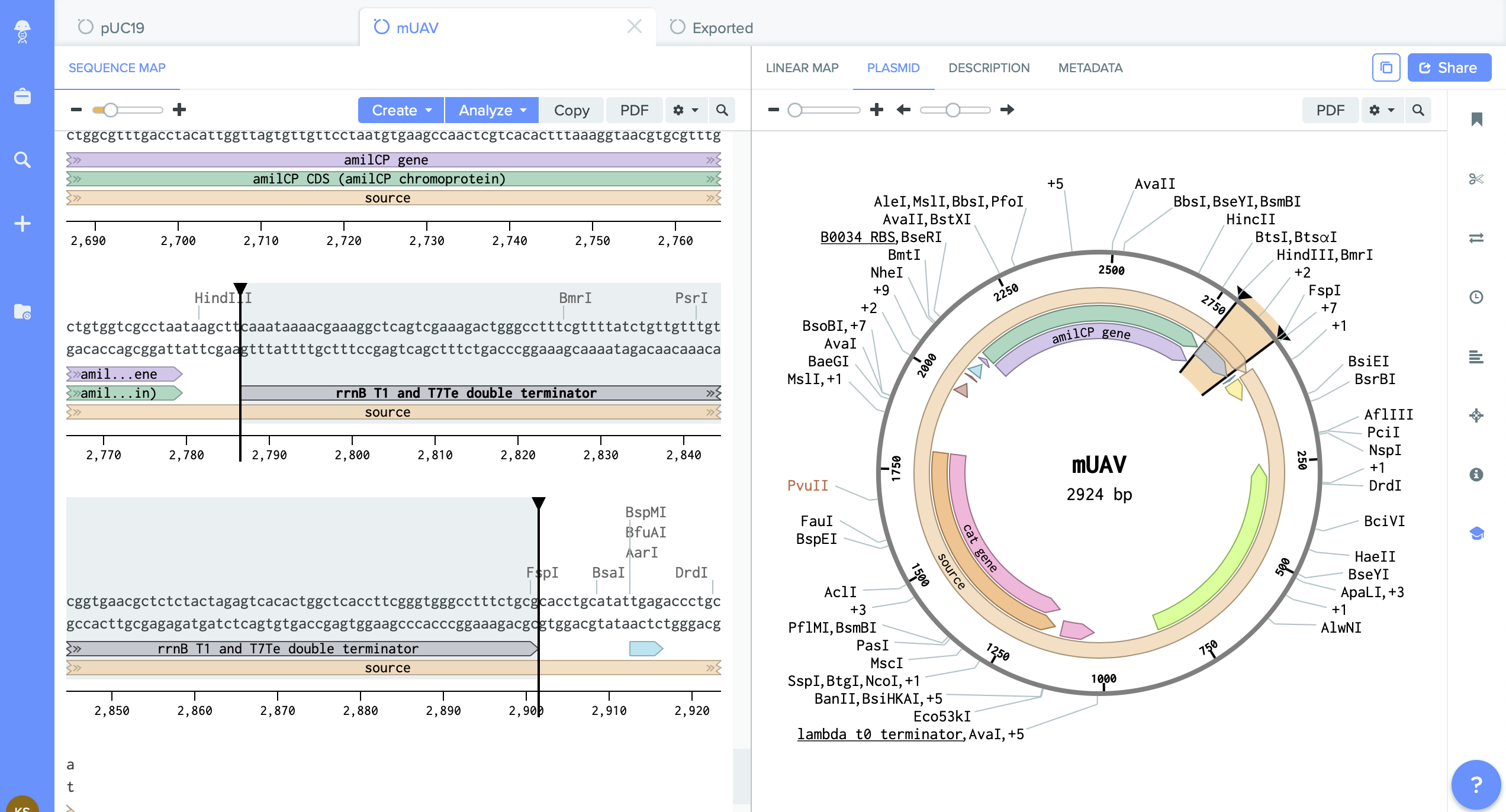
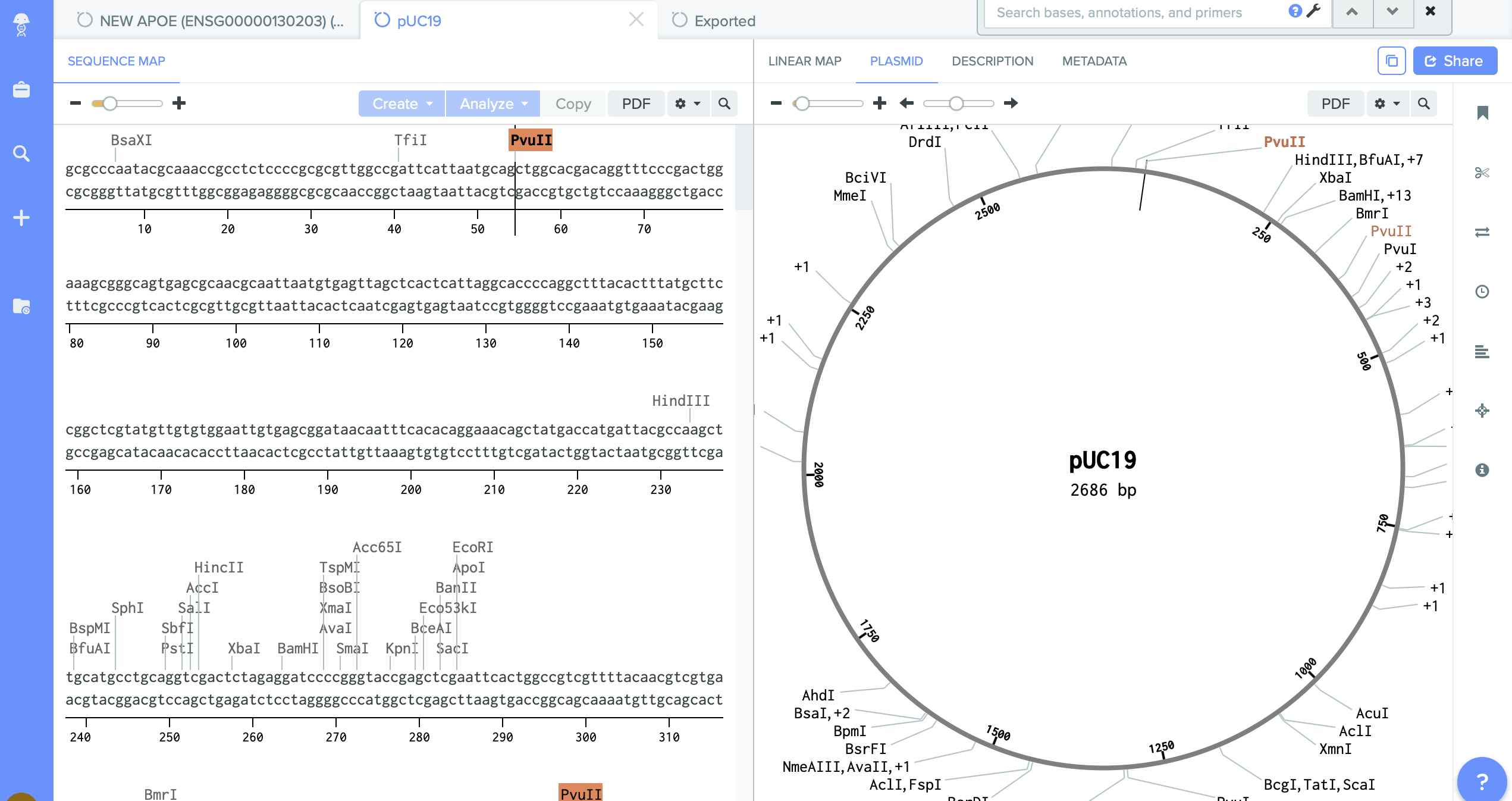
2) pUC19 - empty backbone - cloning vector.
I identified here the cutting region - restriction enzyme site PVU II.
This this the place where I will do cut. And into the space of cut enzyme I am going to insert changed gene amilCP.
Process of designing primers, and changing the gene codon for color orange to code for pink.
So, it took me some time to figure out why do I need four primers.
1) Outer Forward Primer
Outer Forward Primer: (starts with the RBS site, ands just before mutation of the amilCP.
mUAV: 5' ggtatagtgctagctactagag 3'
pUC19: 5' ctggcacgacaggtttcc 3'
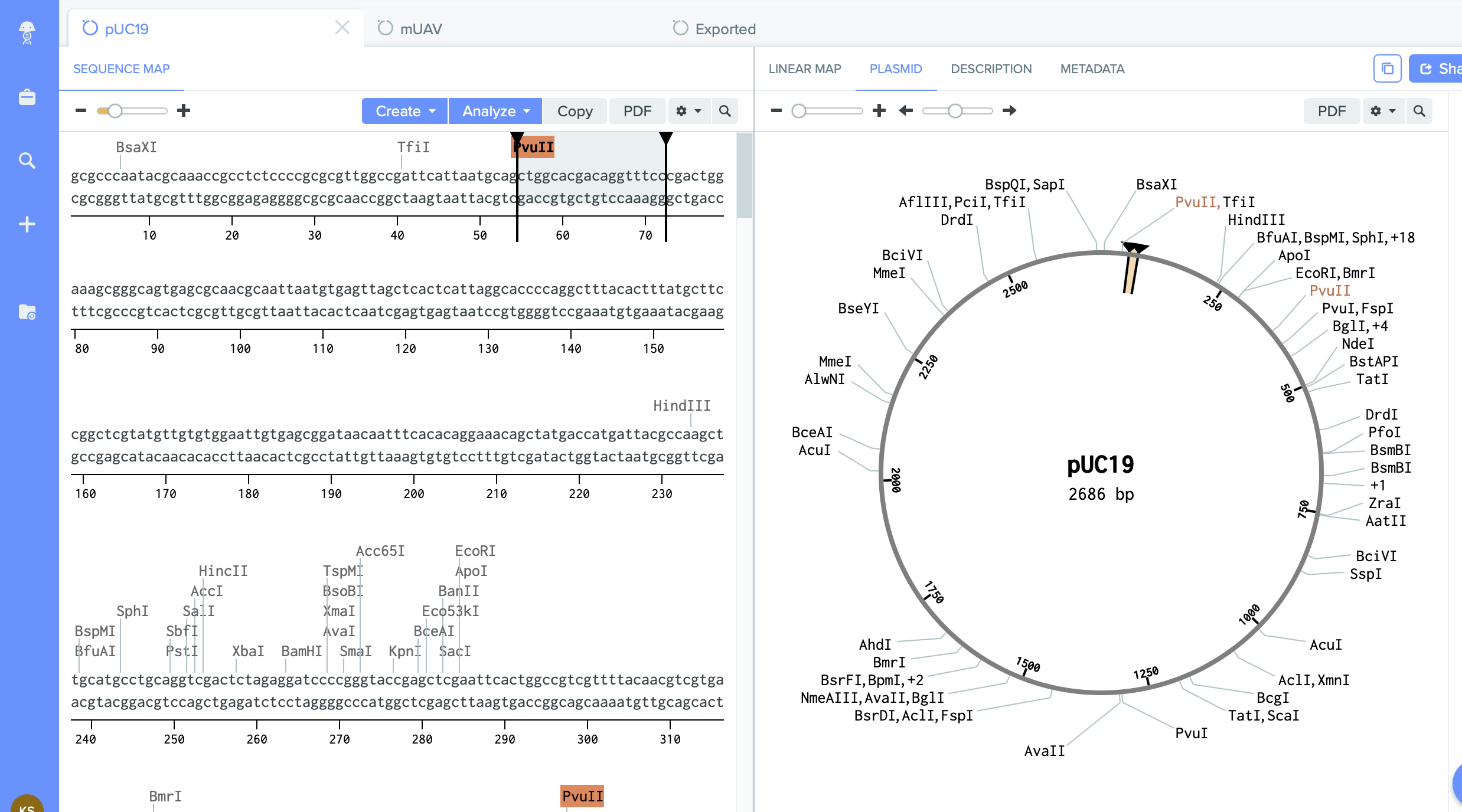
Outer Forward Primer: 5' ctggcacgacaggtttccggtatagtgctagctactagag 3'
length:
Temp:
GC%:
2) Outer Reverse Primer
mUAV: 5' cacctgcatattgagaccctgc 3'
pUC19: 5' ctggcgtaatagcgaagagg 3'
Outer Reverse Primer: 5' cacctgcatattgagaccctgcctggcgtaatagcgaagagg 3'
3) Inner Reverse Primer:
- mUAV: identify the chromophore (CP) mutation region
- Select a 18-24bp region just before the CP mutation region. Right-click and Create Primer (Reverse).
InnerReverse Primer : 5' tggtgataaaatatcccaagca 3'
4) Inner Forward Primer (Mutations):
Instructions
- mUAV: identify the chromophore (CP) mutation region
- Select a 18-24bp region before and after the CP mutation region, as well the the CP mutation region (total length: 18-24 + 6 + 18-24 = 42-54bp!)
- This will be our PCR primer + overhang for Gibson Assembly.
- Copy this sequence to a text editor. Now, use the table you made above to choose which color variant you wish to express.
- You can have multiple colors together! each E.coli cell will only get one plasmid and express it, but we will have many cells and a variety of colors. You can also order different mixes. Go wild.
- To make a mutation library, you have two options:
- Prepare a bunch of primers, that will be synthesized seperately and you will later mix them together using the robot.
- Use degenerate bases, where a single letter (e.g. H) means it could be a number of different bases (e.g. A/C/T).
- For example, if we synthesize the sequence HTGAA, we will get a mixture of: ATGAA, CTGAA, TGTAA in a single tube.
- Your set of sequences is your Inner Forward Primer
before CP mutation region : 5' tttgcttgggatattttatcacca 3'
after CP mutation region : 5' ggaagcataccattcaccaagtacc 3'
PCR Primer + Overhang to Gibson Assembly
Mutation Library:
1) Light Pink
5' tttgcttgggatattttatcaccaGTTTCTggaagcataccattcaccaagtacc 3'
2) Pink
5' tttgcttgggatattttatcaccaGCTTGTggaagcataccattcaccaagtacc 3'
3) Dark Pink
5' tttgcttgggatattttatcaccaGTTCTTggaagcataccattcaccaagtacc 3'
4) Blue
5' tttgcttgggatattttatcaccaGTTAATggaagcataccattcaccaagtacc 3'
In all of my design I tried to keep the suggested parameters for primers design.
Working with Opentrons.
Working with the code was kind of hard for me, since I have 0 coding experience in Phyton. (Definietely something to work on if working remotely).
I also decided to use for the experiment Primers pre prepared not the ones that I designed:
here is my online notebook
A) Isolate pUC19 backbone by Restriction Digest (~30 minutes)
In this step we digest the pUC19 plasmid with the restriction enzyme PvuII to generate the linear blunt-ended backbone fragment.
B) Amplify mutated amilCP fragments by PCR (~1.5 hours)
In this step we will amplify two fragments of amilCP using PCR and our designed primers from last week. One of the primers contains mismatches which will resolve in mutated fragments that could express a variety of colors.
C) Assemble fragments using Gibson Assembly (~30 minutes)
In this step I need to setup a Gibson Assembly of the fragments we generated in parts A and B.
(I have to adjust the times according to new already preprepared primers)
D) Transform into E.coli cells by heat shock (~1.5 hours)
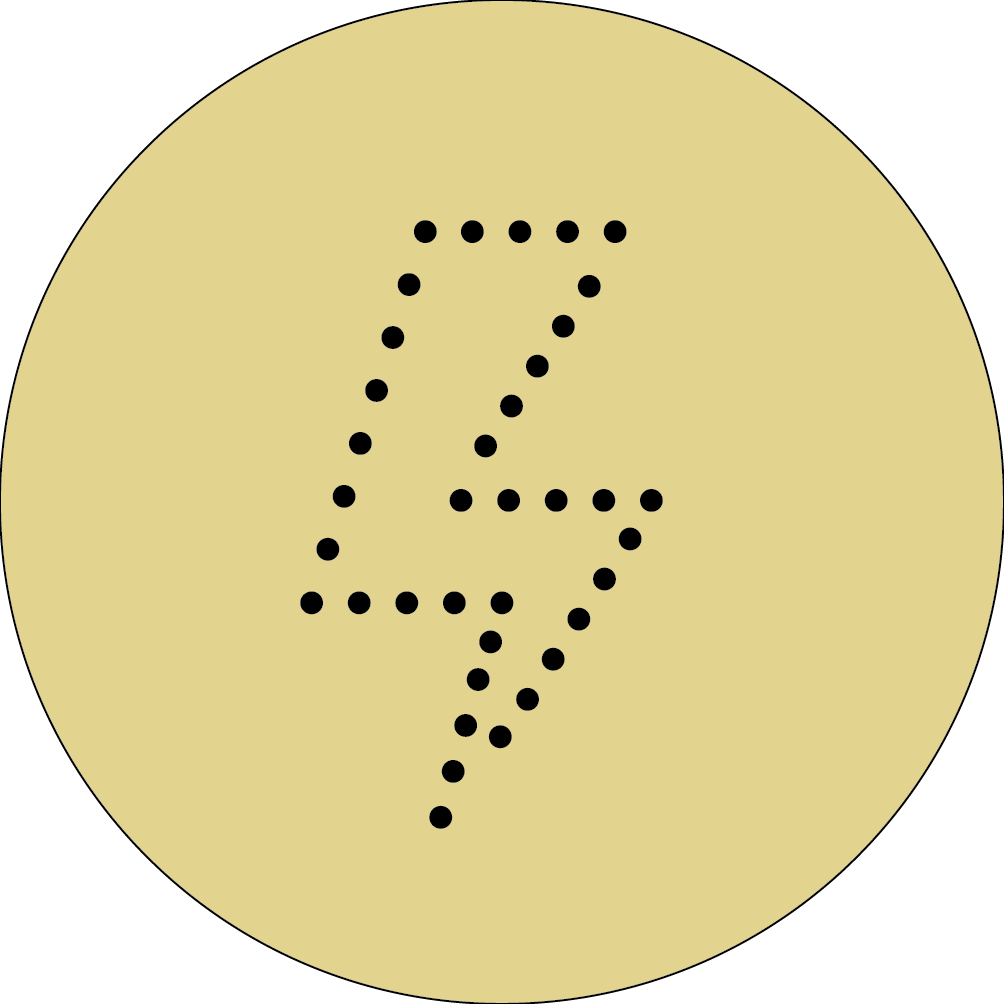
Creating the pattern:
I thought that the best and fastet way would be to do the "drawing" on the plate by strokes.
But I have right now no idea how to code the coordinates.
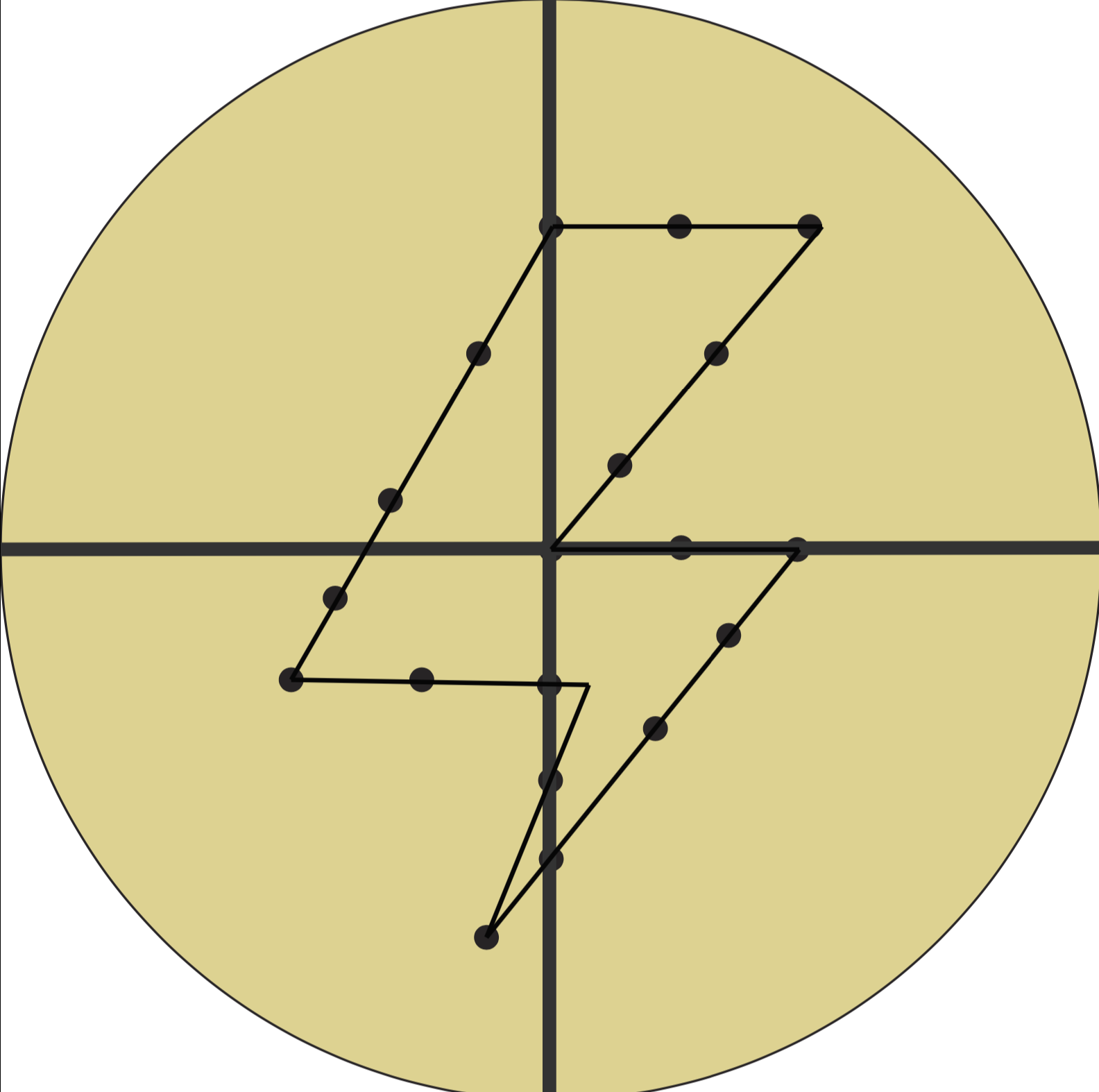
x=horizontal y=vertical
M= move
My coordinates (in centimeters):
- M1 (x-2;y-1) → (x 0, y 2.5)
- M2 (x 0, y 2.5) → (x 2, y 2.5)
- M3 (x 2, y 2.5) → (x 0, y 0)
- M4 (x 0, y 0) → (x 2, y 0)
- M5 (x 2 , y 0) → (x -0.5, y -3)
- M6 (x -0.5, y -3) → (x 0.25, y -1)
- M7 (x 0.25, y -1) → (x -2, y -1)
I also have to assume how many takes of the material the pipette needs to do.
I wanted to do all of that with one tips, and one bacterial color.