Measurement & Imaging Week 5
Part 1: Imaging using FoldScope
I didn't have an access to FoldScope, but I had an access to the microscope.
I also have an access to the sample of :
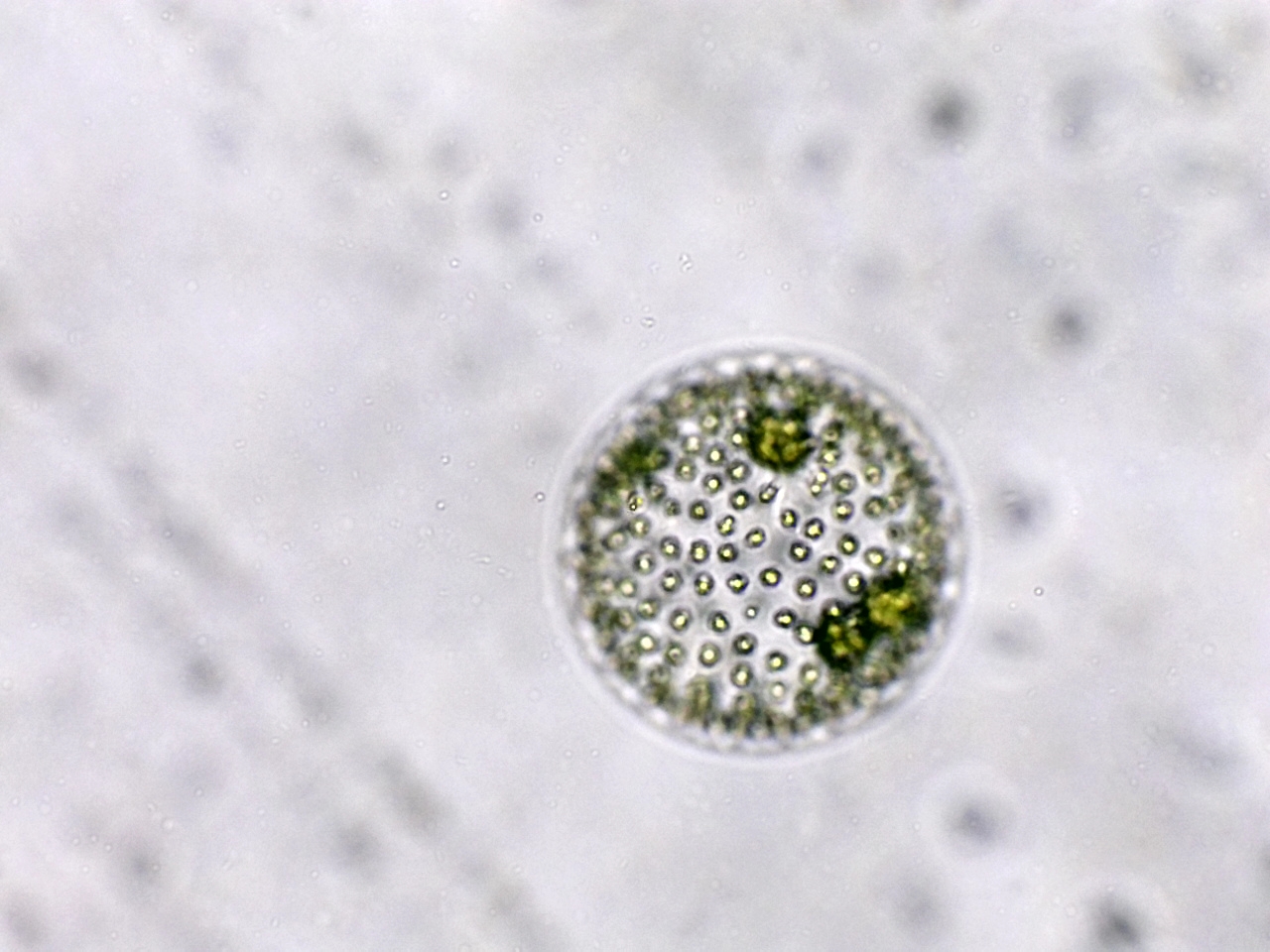
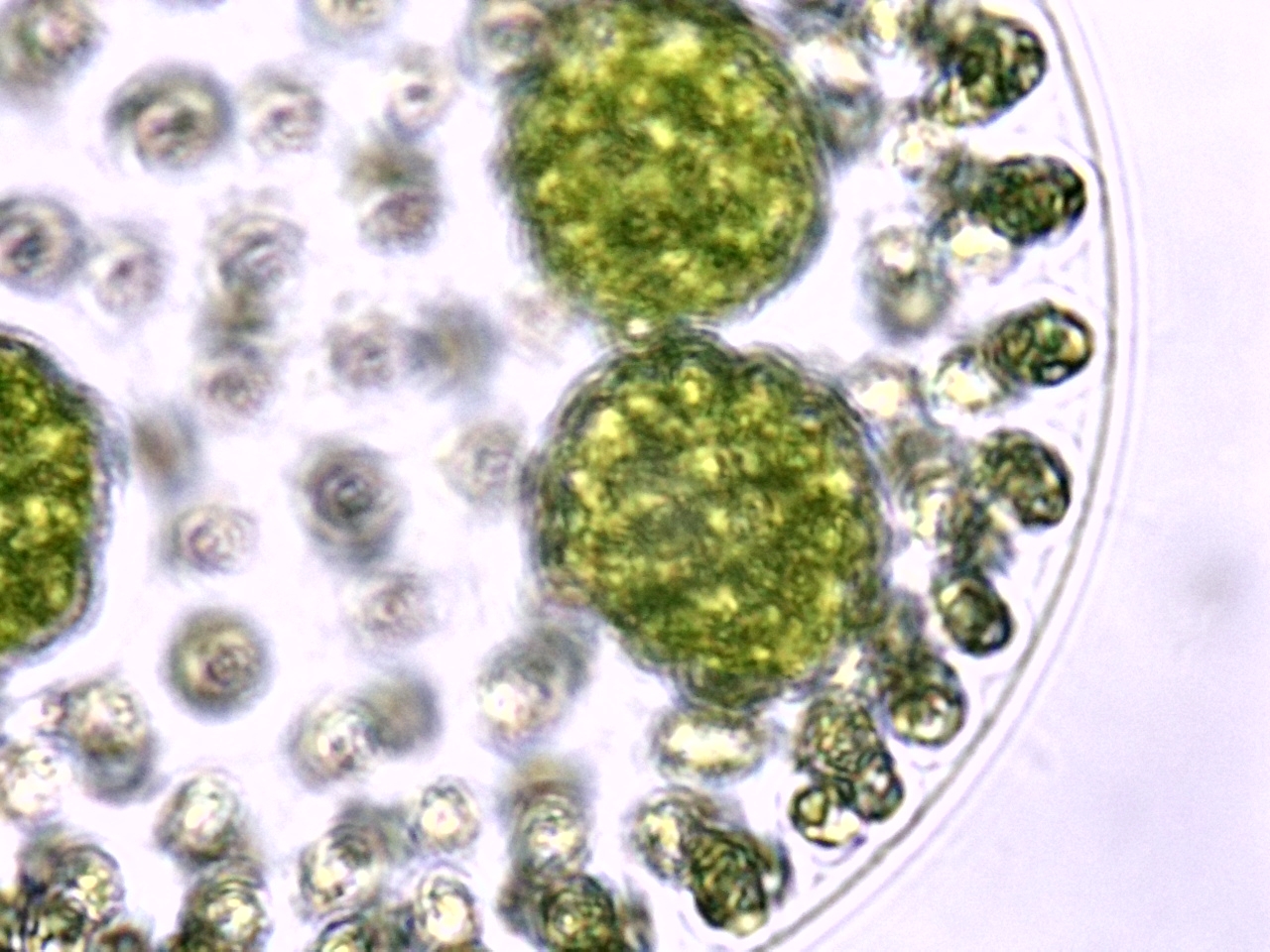
Freshwater green algae
Volvox colonies were first recorded by Dutch microscopist Antonie van Leeuwenhoek in 1700 and are widely studied as a genetic model of morphogenesis (how organisms develop specialized cells and tissues). Volvox also exhibit differentiation between somatic (non-sex cells) and reproductive cells, a phenomenon considered by some biologists to be significant in tracing the evolution of higher animals from microorganisms.
Part 2: Design of smFISH / Spatial Sequencing Assay (Computational)
As detailed in the lecture, most in situ RNA measurement technologies are focused on targeted strategies, which use probes to detect one or more RNA targets. For this part of the homework, you will design a FISH or spatial transcriptomics analysis assay. Design of FISH and RNA capture probes is similar to designing PCR probes.
- Choose a problem. As described in the lecture, a “perfect” measurement experiment can mean a lot of things, but in this case, a perfect assay is one that’s well suited to your biological question. So first, you’ll need to think of an RNA imaging application. Describe why imaging measurement is useful for this problem.
- Pick your target genes. What genes would be relevant to this problem? You can choose any number of genes. Here are some typical examples:
- Diagnostic Assay – Pick one or more genes that are relevant to a disease.
- Synthetic Biology – Pick one or more genes that are involved in a synthetic biology circuit or engineering problem. These could be synthetic genes or endogenous genes, depending on the application. Are you engineering an RNA computer? A synthetic organ-on-chip or organoid?
- What is the assay? Would you use multi-color fluorescence? Multiple cycles? Barcoding? Describe why this readout approach is a good fit.
- Pick one gene and design the homology domains (target sequence domains) for 5 probes.
- Download the RNA sequence of your gene from the relevant database (e.g., FASTA from Refseq). Make sure you are using the mRNA sequence (no introns). Depending on how well annotated your organism and target gene is, you may have to manually process the sequence a little.
- Open the Colab Notebook, make a personal copy (File > Save a Copy) and follow the instructions.
- Pick 5 random sequences, each 25 bases long. Use the “Manual Probe Analyzer” section to find the melting temperature and secondary structure of each of your 5 randomly chosen probe sequences. Present the results & discuss – are these probes good? Are they likely to work well together (e.g., bind under the same conditions)?
- Enter the full target gene sequence in the “PROBE-O-MATIC” designer, as well as a target length, thresholds for min/max temperature, and thresholds for min/max secondary structure. Then run the script.
- Play with the settings and run the script several times. Discuss how the probe design problem is constrained, since length, melting temp, and secondary structure can be related. Did you get enough probes? Note, the script output contains all k-mers within the parameters, with the first base number on the target. However, you’ll want non-overlapping probes in your real design, so pay attention to which probes would be overlapping on the target.
- BLAST one of your probes to see how specific it will be to your target. Does it have hits against other genes? If so, what is the melting temperature against the highest off-target hit (NUPACK is good for this using a 2-strand melting model)? Is this a good probe?