Designing Synthetic Biology Systems. Week 2
This week we had to assignments, one was to extract DNA from our saliva, and the second assignement was to design a mutation in any gene of a mammalian or a plant.
Early considerations
I was thinking for a while which gene to choose, and what procedure to perform. I was considering genes which are related to longevity, cancer, and enhancing cognitive functions.
One of the genes I was considering was Growth Hormonr gene (GH). According to research there is a strong indication that in mice inhibiting the secretion of GH/IGF-1 leads to longer and cancer free life. The way this gene works in humans is not enteirely clear.
I chose to introduce STOP codon in the sequence dedicated to creating GH
This gene is located on 5th chromosome.
The way this gene works in humans is not enteirely clear. Thay is why after all I decided to focus on different gene.
ApoE
Human ApoE exists as three major isoforms, ApoE2, ApoE3 and ApoE4, which are the products of three alleles at a single gene locus on the long arm of chromosome 19.
These isoforms differ structurally by two amino acid substitutions at residues 112 and 158: ApoE2 (Cys112, Cys158), ApoE3 (Cys112, Arg158), and ApoE4 (Arg112, Arg158)
Human ApoE gene is composed of four exons interrupted by three introns.
The ApoE2, ApoE3 and ApoE4 isoforms are coded by ε2, ε3 and ε4 alleles of the ApoE gene, respectively.
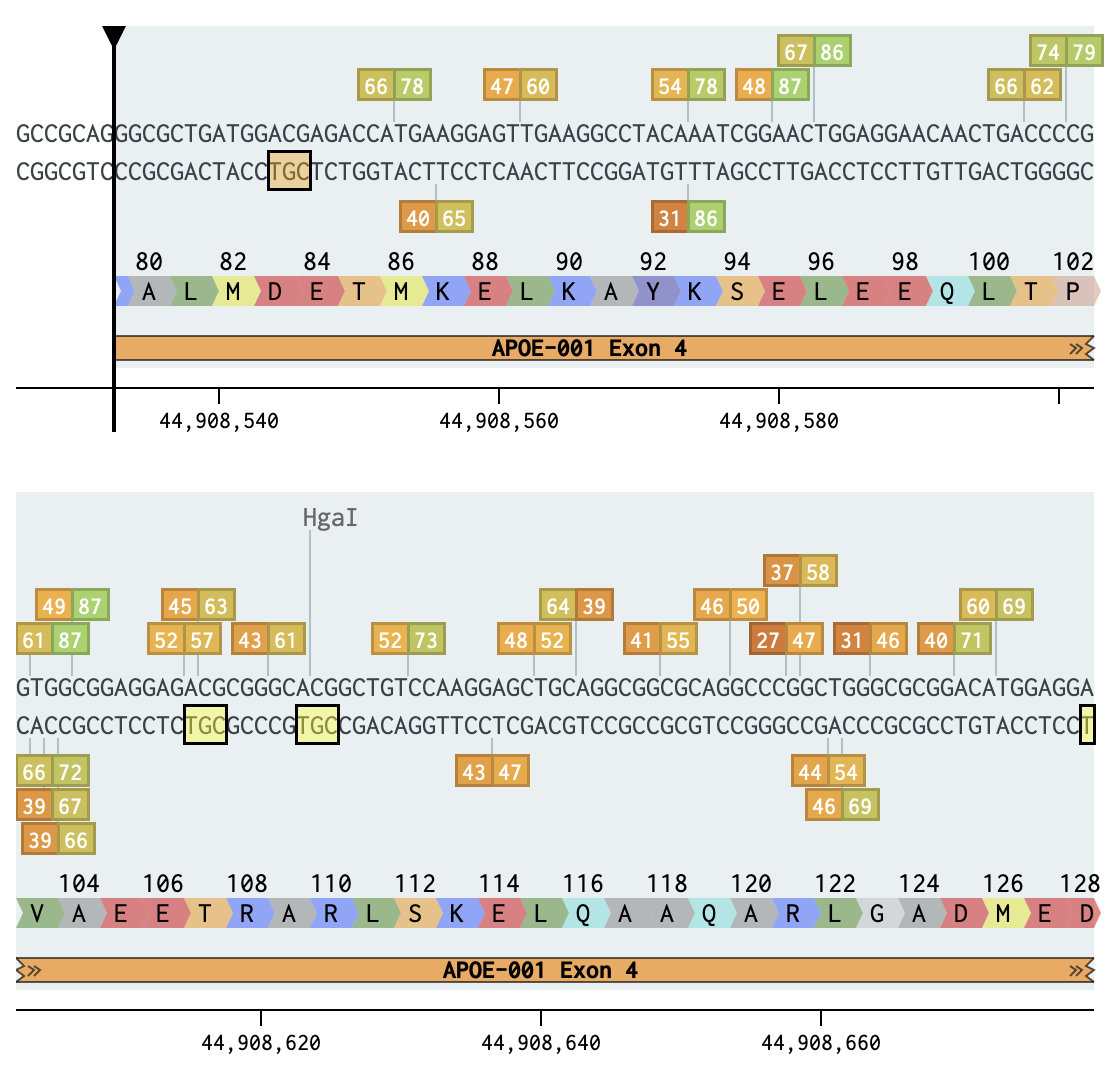
The isoforms differ from each other in two amino acid substitutions at residues 112 and 158 resulting from C→T or T→C point mutation in exon 4 as indicated.
amino acids coding for Cys (Cysteine) are: TGT TGC
amino acids coding for Arg (Arganine) are: CGT CGC CGA CGG
For this assignment we were recommended to use Benchiling, I have never used this tool, and my previous experience with designing on a molecular scale was very limited. I designed a few plasmids some time ago, but I have never used CRISPR/cas9 technique.
I chose genome of GRCh38 (hg38, Homo sapiens) for this assignment.
The genome I chose is (from my investigation) the most common variant - meaning ApoE4 with Arg112 & Arg158. Ther is no Cysteine on the whole Exon4.
I decided to edit first site of Arg and change it to favorable version of Cysteine. Which significantly reduces the possibility of Alzhaimer disease.
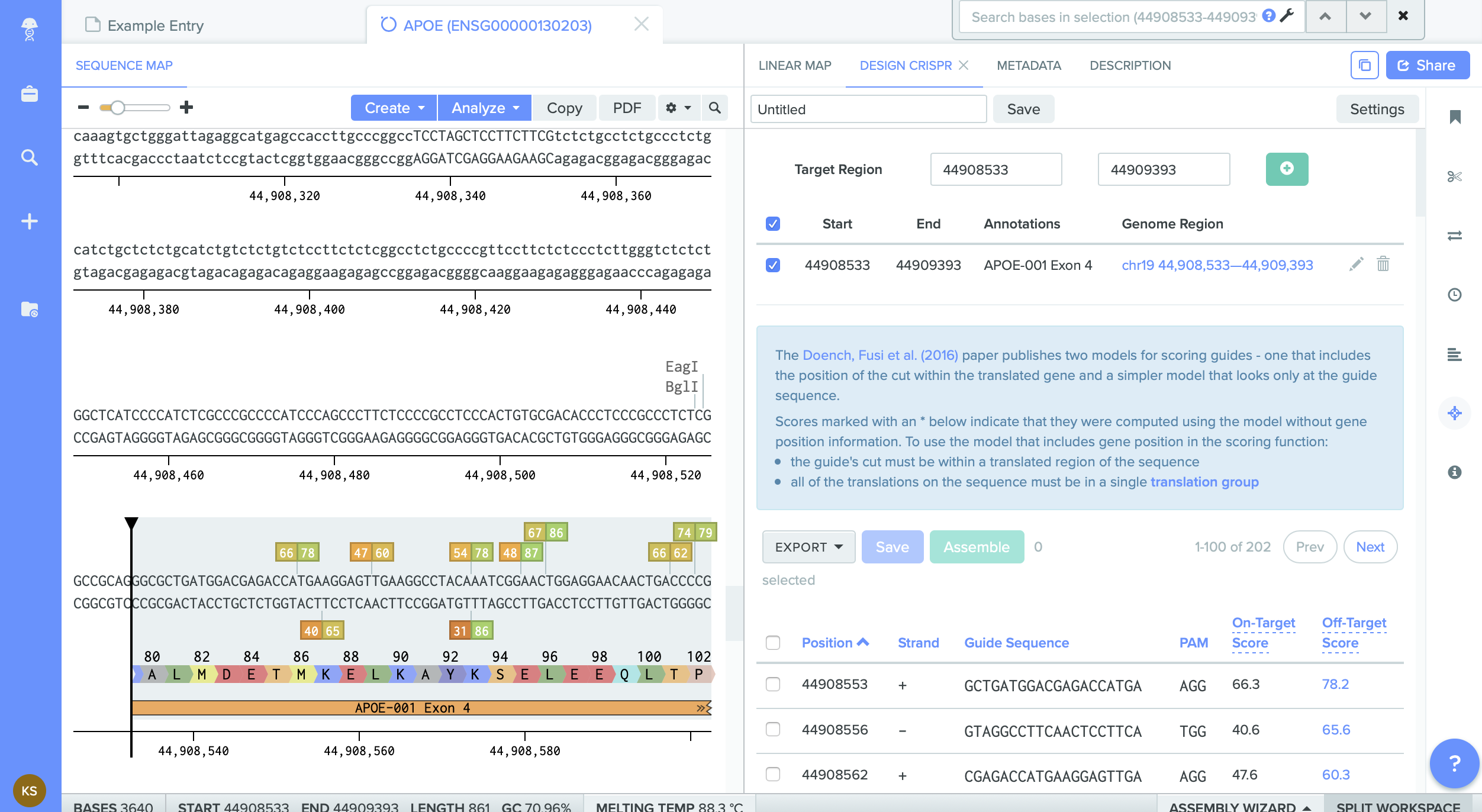
The problem is that in the location I didnt find any Arganine combination.
I am trying to figure out what am I doing wrong... I found some combinations of CGT... but in the Benchling they didn't produce Arg at this location.
I tried to introduce a cut at this location.
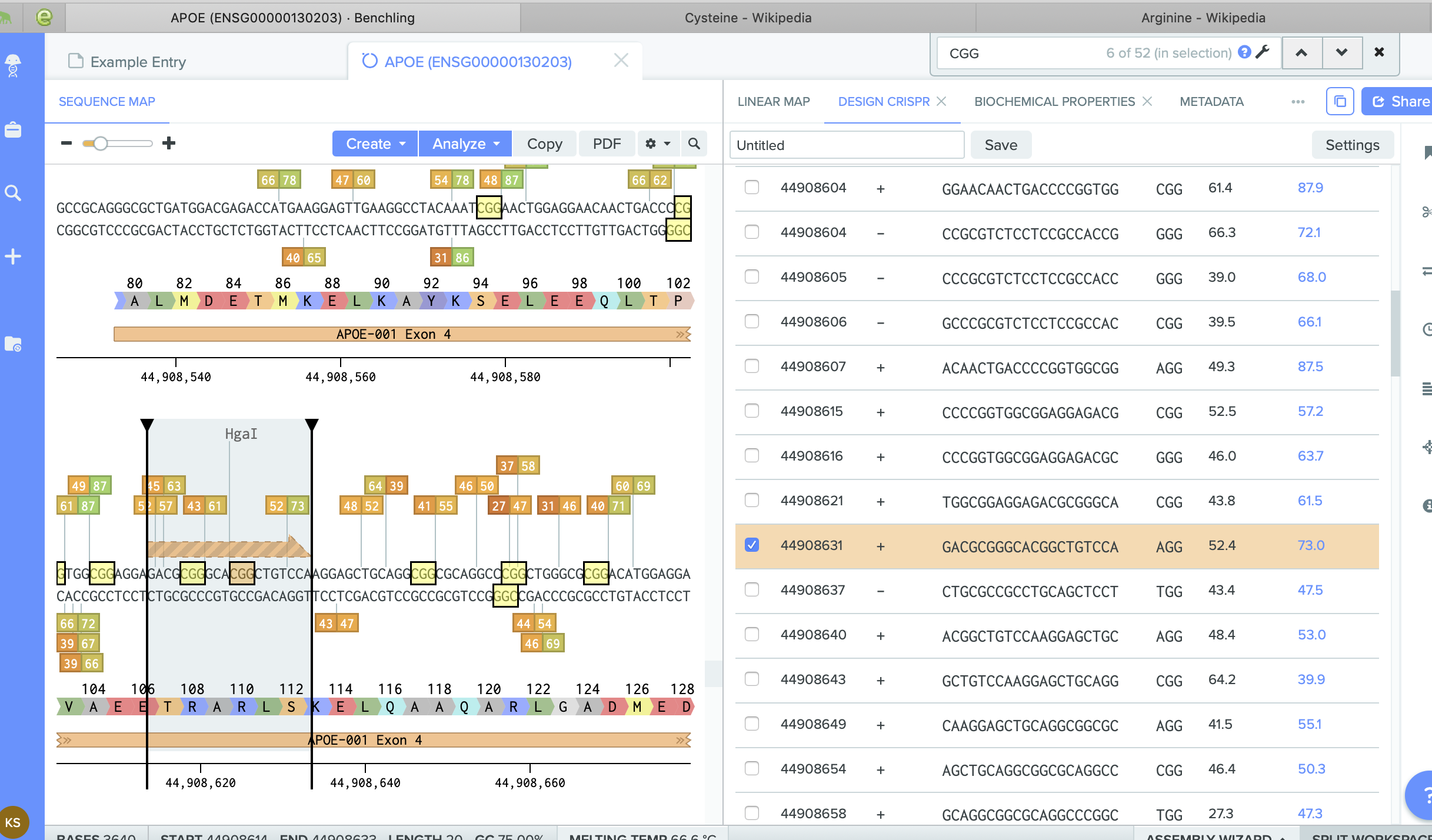
And I got lost with all of that...
I need to re-watch the recitation videos on zoom and have a take two on that.
Sources