Bio-production . Week 7
Part1 : Tele-Experiment Homework
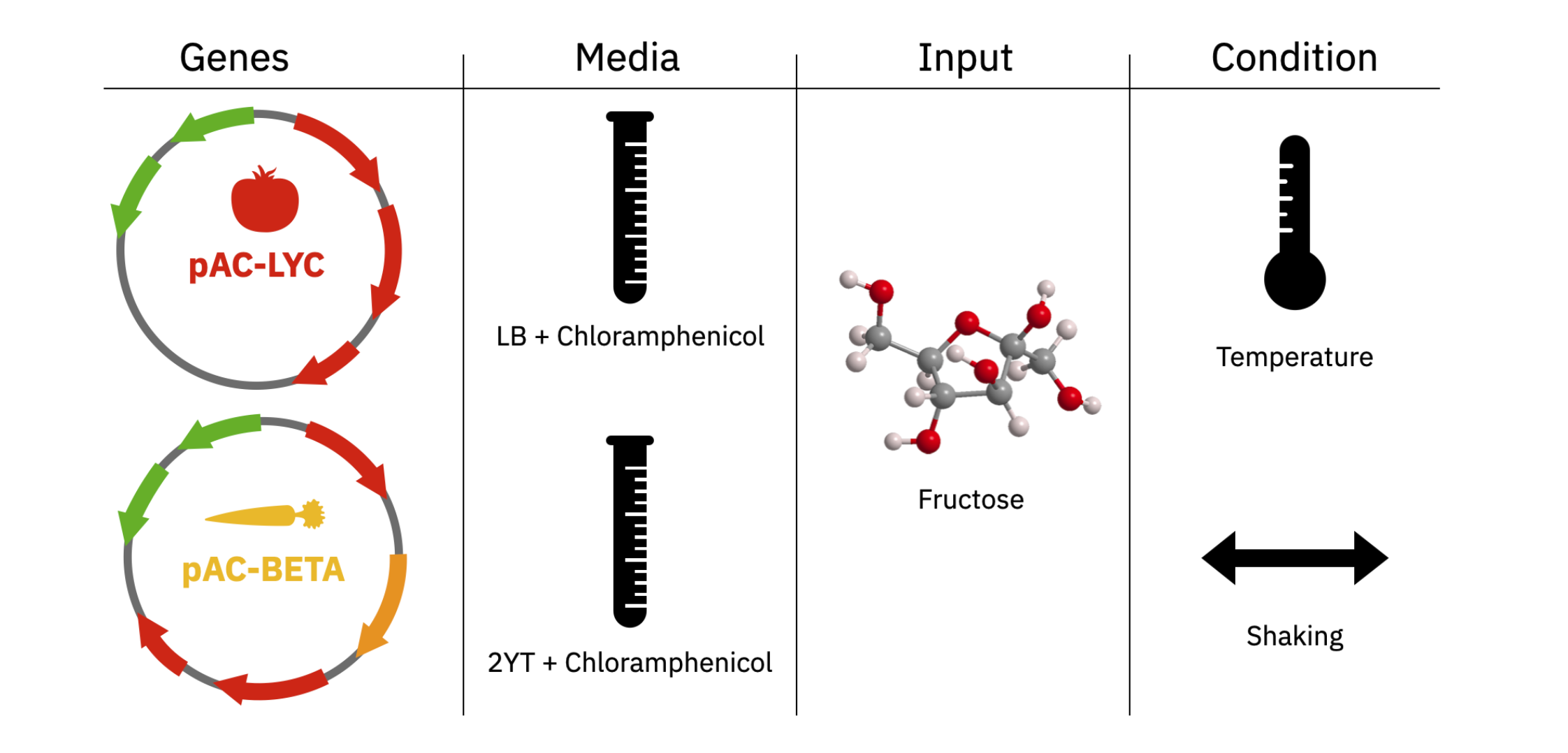
This week we were asked to design and run an experiment to understand how different factors influence bioproduction.
The available factors are genes (pAC-LYC, pAC-BETA), media (LB, 2YT), concentration of precursor (fructose), temperature, and shaking.
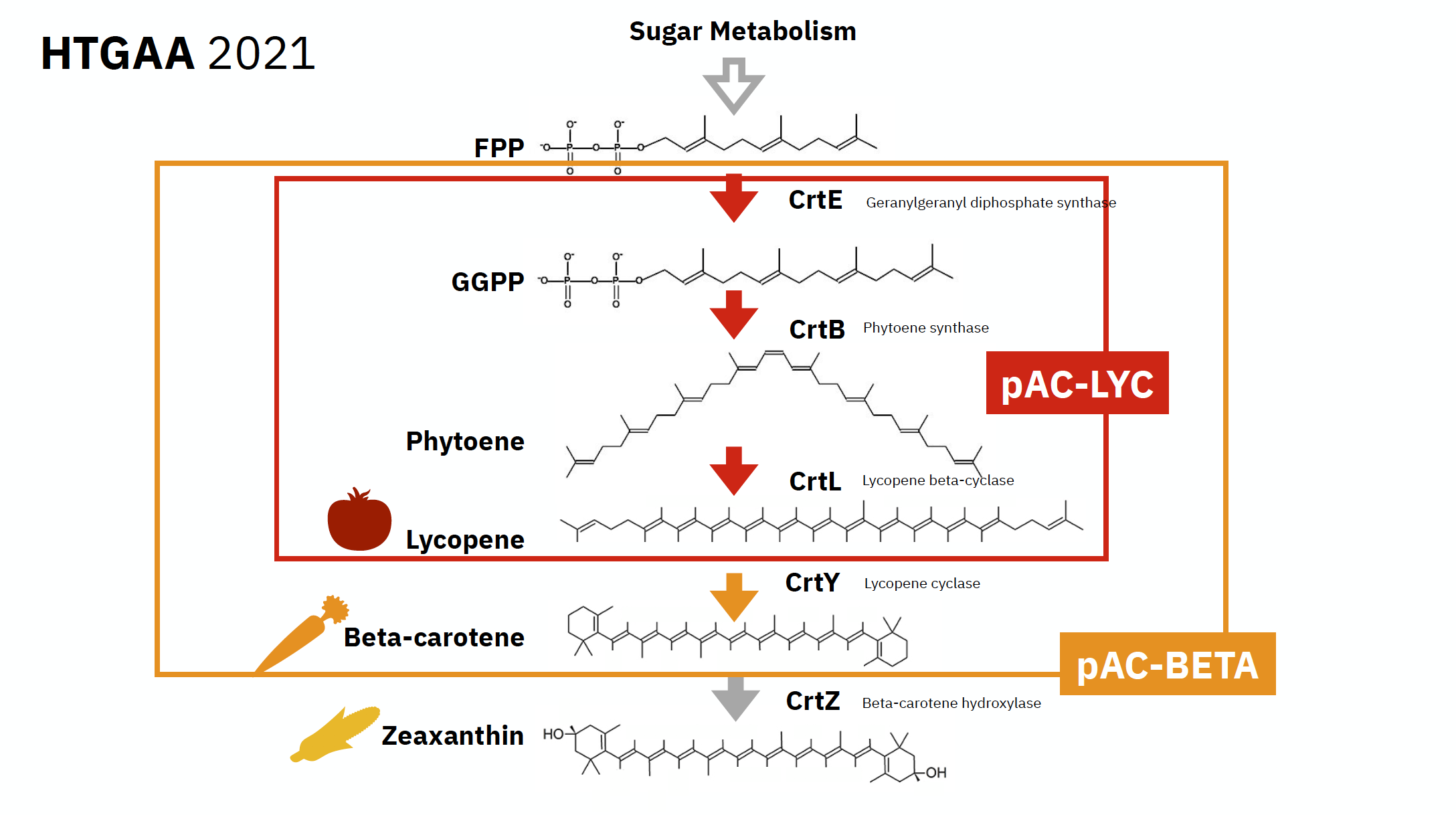
For the class we were focusing on Lycopene
Lycopene has a bright red colour. It is non-provitamin A carotenoid hydrocarbon found in tomatoes and other red fruits, and vegetables (carrots, watermelons, grapefruits, papayas).
Below is a schemata of Lycopene metabolism, showing the pathway of creation on this enzyme:
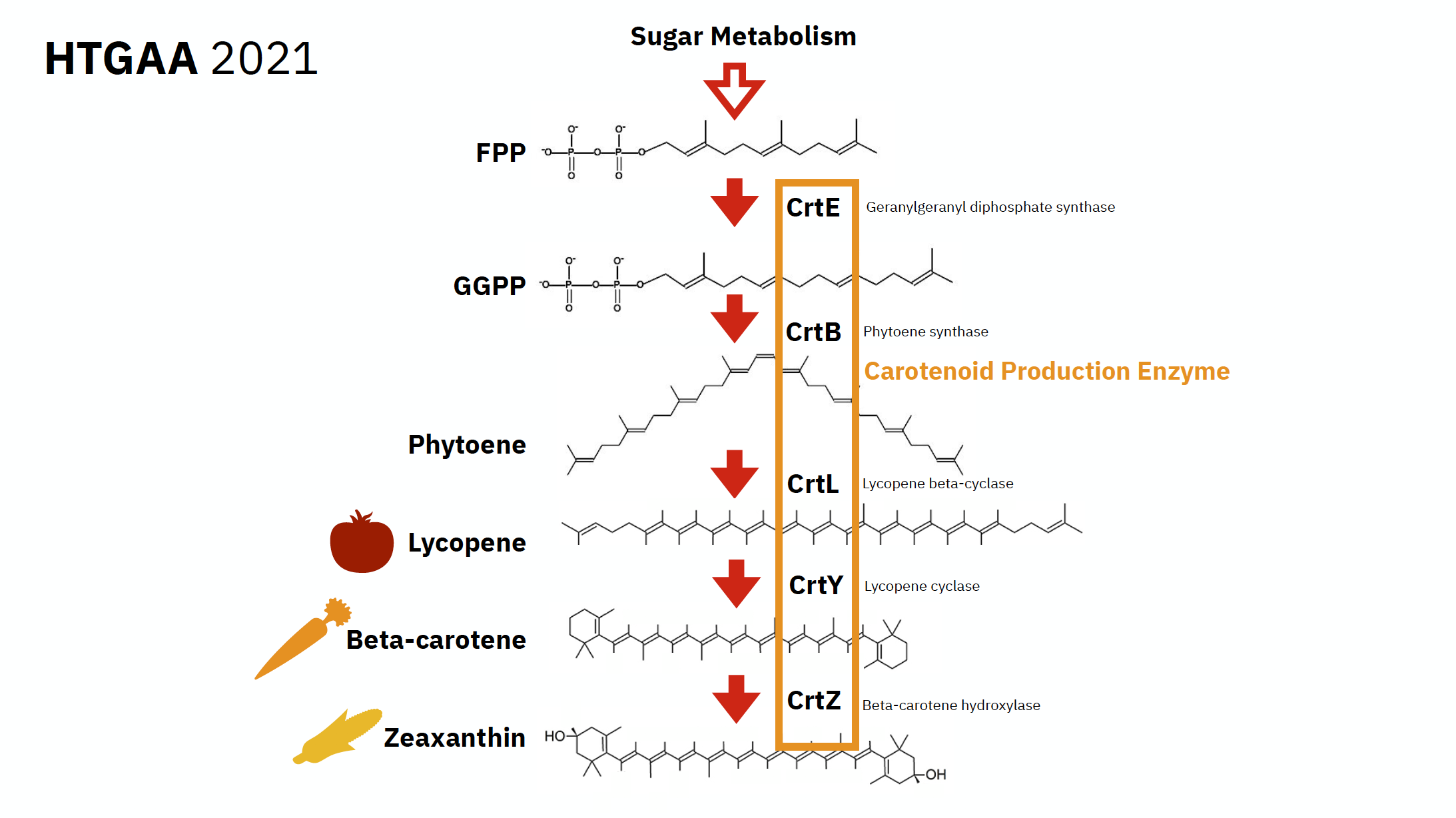
Some notions:
- Carotenoids- Carotenoids are isoprenoid molecules that are synthesized by all photosynthetic organisms and also by some fungi and nonphotosynthetic bacteria. In plants, they participate in photosynthetic light harvesting and protection against light stress. In addition, carotenoids accumulate to large levels as storage metabolites in chromoplasts of flowers, fruits, and taproots.
Carotenoids are also essential to animals, which, however, are unable to synthesise them de novo, and therefore must rely on dietary sources of carotenoids. b-Carotene is the main dietary precursor of vitamin A and therefore also referred to as provitamin A. Vitamin A deficiency in humans represents a global health problem affecting approximately one-third of the countries of the world (Mayer et al., 2008). Presumably due to their antioxidant activity, b-carotene and other carotenoid species also exert protective effects against cardiovascular diseases, certain cancers, and agingrelated diseases (Collins, 1999).
Media:
- LB + Chloramphenicol - Lysogeny Broth and the antibiotic
- 2YT + Chloramphenicol - Trypton , Yeast extract , NaCl & antibiotic
- Plastid - (plant plasmids?)
- fruit chromoplasts - are plastids, heterogeneous organelles responsible for pigment synthesis and storage in specific photosynthetic eukaryotes.[1] It is thought that like all other plastids including chloroplasts and leucoplasts they are descended from symbiotic prokaryotes.
Chromoplasts are found in fruits, flowers, roots, and stressed and aging leaves, and are responsible for their distinctive colors. This is always associated with a massive increase in the accumulation of carotenoid pigments. The conversion of chloroplasts to chromoplasts in ripening is a classic example.
They are generally found in mature tissues and are derived from preexisting mature plastids. Fruits and flowers are the most common structures for the biosynthesis of carotenoids, although other reactions occur there as well including the synthesis of sugars, starches, lipids, aromatic compounds, vitamins, and hormones.[3] The DNA in chloroplasts and chromoplasts is identical.[2] One subtle difference in DNA was found after a liquid chromatography analysis of tomato chromoplasts was conducted, revealing increased cytosine methylation.
- Carotenoids- Carotenoids are isoprenoid molecules that are synthesized by all photosynthetic organisms and also by some fungi and nonphotosynthetic bacteria. In plants, they participate in photosynthetic light harvesting and protection against light stress. In addition, carotenoids accumulate to large levels as storage metabolites in chromoplasts of flowers, fruits, and taproots.
I found an interesting article from 2016 describing an enhancement of lycopene (I attached it at the bottom).
EXPERIMENT
For the experiment were asked to take into consideration the following parameters/factors:
Genes pAC-LYC, pAC-BETA, media (LB Chloramphenicol & 2YT Chloramphenicol), concentration of precursor (fructose), temperature and shaking.
I decided to check how higher input of precursor (fructose) can affect the final product.
concentration of the final product with the same amount of genes.... (?) - actually does it affect anything?
What do you want to compare and why (look at the resources on the website for more ideas)?
I would like to check if there is a possibility to produce more of
Hypothesis
I assume that with more fructose I will have a higer growth, which will lead to more lycopene or beta-caroten.
Control
- Positive Control - added 100 mL fructose ( I will have a bit of the product)
- Negative Control - no fructose added (I will have no product)
- Added 500 ml of fructose (I will have a lot of the product)
- How many replication do you have per condition ?
If I was doing this experiment I would do it two times.
B) Create the code for the tele-experiment on the robot.
Code for extra fructose add on:
metadata = { 'protocolName': 'HTGAA Remote: Bio-production', 'author': 'HTGAA', 'source': 'HTGAA Remote Cloning', 'apiLevel': '2.9' } def run(protocol): ############################################################################## ### Load labware, modules and pipettes ############################################################################## # Tips tips_300ul = protocol.load_labware('opentrons_96_tiprack_300ul', 1, 'Opentrons 300uL Tips') # Tube rack tube_rack = protocol.load_labware('opentrons_10_tuberack_falcon_4x50ml_6x15ml_conical', 3, 'Tube Rack') # Modules #temperature_module = protocol.load_module('temperature module gen2', 9) #thermocycler_module = protocol.load_module('thermocycler') # Incubator incubator = protocol.load_labware('htgaa_24_shaker', 9, label='Incubator') # Pipettes pipette_300ul = protocol.load_instrument("p300_single", "right", tip_racks=[tips_300ul]) ############################################################################## ### Configure starting tips ############################################################################## pipette_300ul.starting_tip = tips_300ul.well('A8') ## CHANGE ME # Test it out by picking and disposing a tip pipette_300ul.pick_up_tip() # Continue only if this step worked! protocol.pause() # Drop tip pipette_300ul.drop_tip() ############################################################################## ### Reagent Configure (Do not change this!) ############################################################################## bacteria_lyc = tube_rack['B2'].bottom(z=10) bacteria_beta = tube_rack['A2'].bottom(z=10) media_LB = tube_rack['A3'].top(z=-60) media_2YT = tube_rack['A4'].top(z=-60) fructose = tube_rack['B3'].top(z=-60) ############################################################################## ### Set tube destination on the Shaker ############################################################################## destination_tube = incubator['A1'].top(z=-40) ############################################################################## ### Add Media ############################################################################## # Dispense 2.5ml of media pipette_300ul.pick_up_tip() for i in range(8): pipette_300ul.aspirate(30, media_LB) ## Change this! pipette_300ul.dispense(30, destination_tube) ## Change this! pipette_300ul.blow_out() pipette_300ul.aspirate(100, media_LB) pipette_300ul.dispense(100, destination_tube) pipette_300ul.blow_out() pipette_300ul.drop_tip() ############################################################################## ### Add Microbes ############################################################################## pipette_300ul.pick_up_tip() pipette_300ul.aspirate(30, bacteria_lyc) ## Change this! pipette_300ul.dispense(30, destination_tube) ## Change this! pipette_300ul.mix(5, 300, destination_tube) pipette_300ul.blow_out() pipette_300ul.drop_tip() ############################################################################## ### Add Fructose ############################################################################## pipette_300ul.pick_up_tip() pipette_300ul.aspirate(250, fructose) ## Change this! pipette_300ul.dispense(250, destination_tube) ## Change this! pipette_300ul.mix(5, 300, destination_tube) pipette_300ul.aspirate(250, fructose) ## pipette_300ul.dispense(250, destination_tube) ## pipette_300ul.blow_out() pipette_300ul.drop_tip() ############################################################################## ### Activate! ############################################################################## #temperature_module.set_temperature(37) ## SHAKE IT!! ## using arduino
I also tried to play a little bit with the code here: ......
C) Run the simulation to make sure that there is no syntax error
👍🏼 - in the simulation there was no error
Part 2: Identify a cool gene (related to your final project)
For indentyfing cool gene I decided to go with:
Since I have been thinking about doing "electric E.coli", I was thinking of focusing on the gene of G. sulfurreducens, but l found out thanks to friend from the class Weiting that the iGEM group did a great job with this gene. Here is their work: link.
There is bacteria with similar properities is Shewanella oneidensis MR-1.
Part 3: If you have the capability of Gingko Bioworks foundry...
Write a short paragraph responding to "If you have the capability of Gingko Bioworks foundry, what would you do and why?" These could be COVID19 related projects or broader synthetic biology projects. Ginkgo Bioworks is taking proposals to leverage the use of their platform to support technical projects; how might you leverage Ginkgo’s technical platform to support your project? Write a description of how you would utilize their platform. What tools and capacities would you use? How would you use them? How would the use of Ginkgo’s platform accelerate or increase the technical capabilities of your project?
What is cool about Gingko:
- Possibility to synthesis, screen, and improvement thousand enzyme libraries.
- designing organisms - automation, scale, ability to collect and analyze a lot of data.
- using the Foundry approach
- scaling up and optimisation of processes such as fermentation. (automation of Sartorius ambr250 mL disposable reactor system - possibility to run hundreds of bioreactors simultaneously)
How I would use Gingko capacities:
Bibliography:
Lycopene
1) https://pubmed.ncbi.nlm.nih.gov/19587100/
2) https://pubmed.ncbi.nlm.nih.gov/33073979/
3) Regulation of carotenoid metabolism in tomato - https://pubmed.ncbi.nlm.nih.gov/25578270/
4) Analysis and expression of the carotenoid biosynthesis genes from Deinococcus wulumuqiensis R12 in engineered Escherichia coli (very interesting that extremophile such as Deinococcus can contribute to the production of carotenoids)- link
E.coli